AnchorQuery™ |
||||||||||||
|
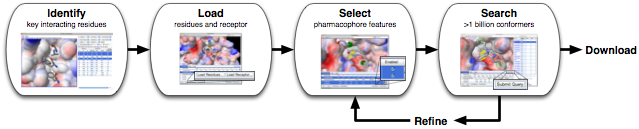
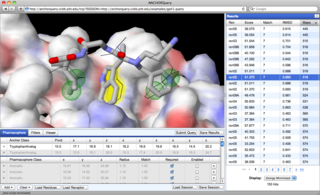
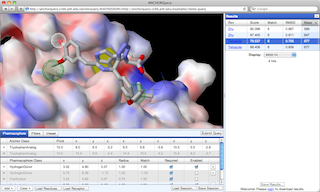
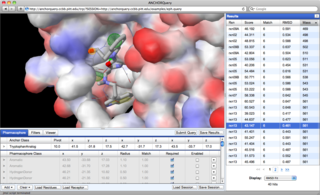
AnchorQuery™ is a specialized
pharmacophore
search technology that brings interactive virtual screening of novel protein-protein inhibitors to the desktop.
Try one of the interactive examples or read the Getting Started
guide in the User Manual to search for a custom pharmacophore.
Statistics7 anchor analogs (F,L,W,V,Y,D,E)31,139,128 compounds 2,002,551,771 conformers Search Anchor Biased Library Library Information
Extract PPI inhibitor starting point pharmacophores from PPI structure with PocketQuery.
Interactive Examples
Background
AnchorQuery™ leverages the concept
of anchors, amino acid residues that bury a large amount of
solvent accessible surface area at the protein-protein interface. Every
compound in our multi-component reaction (MCR) accessible
virtual library contains an anchor analog, a functional group
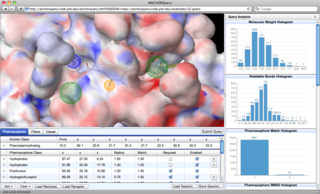
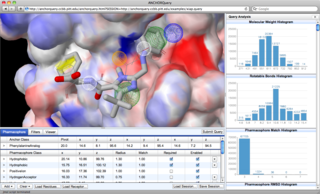
that is a chemical mimic of a specific amino acid. AnchorQuery™ pharmacophore
queries always include an anchor feature in addition to the standard
hydrophobic, ionic, and hydrogen bond donors. All non-anchor features
are stored relative to a coordinate system defined by the anchor in an
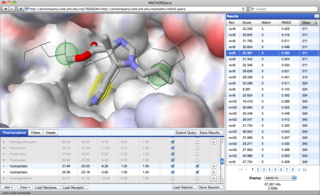
efficient spatial index. Pharmacophore searches scale
relative to the breadth and complexity of the query, not the size of the
database. As a result, full 3D pharmacophore searches can be executed
over millions of explicit conformations in a matter of seconds.
|